Mapping the genome of the bumblebee Bombus terrestris.

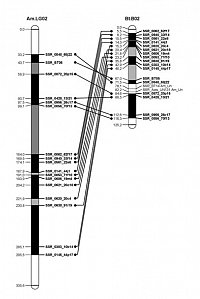
As being an emerging model system, there is a need for the development of genetic and genomic resources for Bombus terrestris. A crucial part is a genetic linkage map. Such a map helps to understand the genetic organization (chromosomes), the recombination rate and enables QTL mapping studies and finally will be a substantial improvement of the genome assembly as genomic scaffolds can be placed and orientated on chromosomes. This further facilitates comparative genomics. During our study we developed in a collaborative work with the research groups of Paul Schmid-Hempel and Regula Schmid-Hempel (ETH Zürich) novel microsatellite (SSR) markers to saturate the B. terrestris genome. This was done by an enrichment and sequencing approach as well as using BAC-end library sequences. We detected 16 linkage groups with a total length of 2047 cM which translates into a recombinbation rate of about 8.2 if a genome size of 250 Mbp is assumed. We also detected large homologies in genome structure compared to Apis mellifera, although both are about 100 million years apart.
- Stolle E, Wilfert P, Schmid-Hempel R, Schmid-Hempel P, Kube M, Reinhardt R, Moritz RFA (2011): A second generation genetic map of the bumblebee Bombus terrestris (Linnaeus, 1758) reveals slow genome and chromosome evolution in the Apidae. BMC Genomics 12: 48
- Stolle E, Rohde M, Vautrin D, Solignac M, Schmid-Hempel P, Schmid-Hempel R, Moritz RFA (2009): Novel microsatellite DNA loci for Bombus terrestris (Linnaeus, 1758). Molecular Ecology Resources 9 (5): 1345–1352
Mapping the genome of the bumblebee Bombus terrestris.
Funded by:



